Introduction
The data set is the mushroom set retrieved from: http://archive.ics.uci.edu/ml/datasets/Mushroom. It consists of 8124 orbs and 23 variables. The classification variable is the “type” either edible (e) or poisonous (p). The remaining 22 variables are the predictors and consist of multiple levels from 1 to 12 each. The data columns will need to be named and are coded in short for each. There are 2480 NA’s found and in one variable “sr” or stalk root. It is decided to use kNN imputation (k=10) to fill these values in with one of the five levels found in this variable.The imputation works for categorical data and thus chosen for this task. It is decided to create dummy variables into numeric values (1,0), but first the veil-type “vt” only has one level and will be removed, also the imputation creates a sr_imp variable and this is also removed. There will be 117 variables, to reduce collinearity the 2 level variables are reduced to 1 variable (fullRank=T). The dependent variable “type” then added back to the revised set as a factor or 2 levels (e,p). The first section will analyze the ANN using the nnet() from the nnet package. The second section will analyze the SVM using the ksvm() from the kernlab package.
library(VIM)
library(caret)
#ANN Libs
library(nnet)
library(RCurl)
library(Metrics)agaricus.lepiota <- read.csv("http://archive.ics.uci.edu/ml/machine-learning-databases/mushroom/agaricus-lepiota.data", header=FALSE, row.names=NULL, na.strings="?")mr <- agaricus.lepiota
#View(mr)
str(mr)## 'data.frame': 8124 obs. of 23 variables:
## $ V1 : Factor w/ 2 levels "e","p": 2 1 1 2 1 1 1 1 2 1 ...
## $ V2 : Factor w/ 6 levels "b","c","f","k",..: 6 6 1 6 6 6 1 1 6 1 ...
## $ V3 : Factor w/ 4 levels "f","g","s","y": 3 3 3 4 3 4 3 4 4 3 ...
## $ V4 : Factor w/ 10 levels "b","c","e","g",..: 5 10 9 9 4 10 9 9 9 10 ...
## $ V5 : Factor w/ 2 levels "f","t": 2 2 2 2 1 2 2 2 2 2 ...
## $ V6 : Factor w/ 9 levels "a","c","f","l",..: 7 1 4 7 6 1 1 4 7 1 ...
## $ V7 : Factor w/ 2 levels "a","f": 2 2 2 2 2 2 2 2 2 2 ...
## $ V8 : Factor w/ 2 levels "c","w": 1 1 1 1 2 1 1 1 1 1 ...
## $ V9 : Factor w/ 2 levels "b","n": 2 1 1 2 1 1 1 1 2 1 ...
## $ V10: Factor w/ 12 levels "b","e","g","h",..: 5 5 6 6 5 6 3 6 8 3 ...
## $ V11: Factor w/ 2 levels "e","t": 1 1 1 1 2 1 1 1 1 1 ...
## $ V12: Factor w/ 4 levels "b","c","e","r": 3 2 2 3 3 2 2 2 3 2 ...
## $ V13: Factor w/ 4 levels "f","k","s","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ V14: Factor w/ 4 levels "f","k","s","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ V15: Factor w/ 9 levels "b","c","e","g",..: 8 8 8 8 8 8 8 8 8 8 ...
## $ V16: Factor w/ 9 levels "b","c","e","g",..: 8 8 8 8 8 8 8 8 8 8 ...
## $ V17: Factor w/ 1 level "p": 1 1 1 1 1 1 1 1 1 1 ...
## $ V18: Factor w/ 4 levels "n","o","w","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ V19: Factor w/ 3 levels "n","o","t": 2 2 2 2 2 2 2 2 2 2 ...
## $ V20: Factor w/ 5 levels "e","f","l","n",..: 5 5 5 5 1 5 5 5 5 5 ...
## $ V21: Factor w/ 9 levels "b","h","k","n",..: 3 4 4 3 4 3 3 4 3 3 ...
## $ V22: Factor w/ 6 levels "a","c","n","s",..: 4 3 3 4 1 3 3 4 5 4 ...
## $ V23: Factor w/ 7 levels "d","g","l","m",..: 6 2 4 6 2 2 4 4 2 4 ...names(mr)[1:23] <- c("type","csh", "csf", "cc", "b", "o", "ga", "gs", "gz", "gc", "ss", "sr", "ssar", "ssbr", "scar", "scbr", "vt", "vc", "rn", "rt","spc","p", "h")
str(mr)## 'data.frame': 8124 obs. of 23 variables:
## $ type: Factor w/ 2 levels "e","p": 2 1 1 2 1 1 1 1 2 1 ...
## $ csh : Factor w/ 6 levels "b","c","f","k",..: 6 6 1 6 6 6 1 1 6 1 ...
## $ csf : Factor w/ 4 levels "f","g","s","y": 3 3 3 4 3 4 3 4 4 3 ...
## $ cc : Factor w/ 10 levels "b","c","e","g",..: 5 10 9 9 4 10 9 9 9 10 ...
## $ b : Factor w/ 2 levels "f","t": 2 2 2 2 1 2 2 2 2 2 ...
## $ o : Factor w/ 9 levels "a","c","f","l",..: 7 1 4 7 6 1 1 4 7 1 ...
## $ ga : Factor w/ 2 levels "a","f": 2 2 2 2 2 2 2 2 2 2 ...
## $ gs : Factor w/ 2 levels "c","w": 1 1 1 1 2 1 1 1 1 1 ...
## $ gz : Factor w/ 2 levels "b","n": 2 1 1 2 1 1 1 1 2 1 ...
## $ gc : Factor w/ 12 levels "b","e","g","h",..: 5 5 6 6 5 6 3 6 8 3 ...
## $ ss : Factor w/ 2 levels "e","t": 1 1 1 1 2 1 1 1 1 1 ...
## $ sr : Factor w/ 4 levels "b","c","e","r": 3 2 2 3 3 2 2 2 3 2 ...
## $ ssar: Factor w/ 4 levels "f","k","s","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ ssbr: Factor w/ 4 levels "f","k","s","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ scar: Factor w/ 9 levels "b","c","e","g",..: 8 8 8 8 8 8 8 8 8 8 ...
## $ scbr: Factor w/ 9 levels "b","c","e","g",..: 8 8 8 8 8 8 8 8 8 8 ...
## $ vt : Factor w/ 1 level "p": 1 1 1 1 1 1 1 1 1 1 ...
## $ vc : Factor w/ 4 levels "n","o","w","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ rn : Factor w/ 3 levels "n","o","t": 2 2 2 2 2 2 2 2 2 2 ...
## $ rt : Factor w/ 5 levels "e","f","l","n",..: 5 5 5 5 1 5 5 5 5 5 ...
## $ spc : Factor w/ 9 levels "b","h","k","n",..: 3 4 4 3 4 3 3 4 3 3 ...
## $ p : Factor w/ 6 levels "a","c","n","s",..: 4 3 3 4 1 3 3 4 5 4 ...
## $ h : Factor w/ 7 levels "d","g","l","m",..: 6 2 4 6 2 2 4 4 2 4 ...summary(mr, maxsum = 12)## type csh csf cc b o ga gs
## e:4208 b: 452 f:2320 b: 168 f:4748 a: 400 a: 210 c:6812
## p:3916 c: 4 g: 4 c: 44 t:3376 c: 192 f:7914 w:1312
## f:3152 s:2556 e:1500 f:2160
## k: 828 y:3244 g:1840 l: 400
## s: 32 n:2284 m: 36
## x:3656 p: 144 n:3528
## r: 16 p: 256
## u: 16 s: 576
## w:1040 y: 576
## y:1072
##
##
## gz gc ss sr ssar ssbr scar scbr
## b:5612 b:1728 e:3516 b :3776 f: 552 f: 600 b: 432 b: 432
## n:2512 e: 96 t:4608 c : 556 k:2372 k:2304 c: 36 c: 36
## g: 752 e :1120 s:5176 s:4936 e: 96 e: 96
## h: 732 r : 192 y: 24 y: 284 g: 576 g: 576
## k: 408 NA's:2480 n: 448 n: 512
## n:1048 o: 192 o: 192
## o: 64 p:1872 p:1872
## p:1492 w:4464 w:4384
## r: 24 y: 8 y: 24
## u: 492
## w:1202
## y: 86
## vt vc rn rt spc p h
## p:8124 n: 96 n: 36 e:2776 b: 48 a: 384 d:3148
## o: 96 o:7488 f: 48 h:1632 c: 340 g:2148
## w:7924 t: 600 l:1296 k:1872 n: 400 l: 832
## y: 8 n: 36 n:1968 s:1248 m: 292
## p:3968 o: 48 v:4040 p:1144
## r: 72 y:1712 u: 368
## u: 48 w: 192
## w:2388
## y: 48
##
##
## summary(mr$sr)## b c e r NA's
## 3776 556 1120 192 2480#####################Prepare data
set.seed(1234)
sr_imp <- kNN(mr, variable = "sr", k=10, impNA=TRUE)
str(sr_imp)## 'data.frame': 8124 obs. of 24 variables:
## $ type : Factor w/ 2 levels "e","p": 2 1 1 2 1 1 1 1 2 1 ...
## $ csh : Factor w/ 6 levels "b","c","f","k",..: 6 6 1 6 6 6 1 1 6 1 ...
## $ csf : Factor w/ 4 levels "f","g","s","y": 3 3 3 4 3 4 3 4 4 3 ...
## $ cc : Factor w/ 10 levels "b","c","e","g",..: 5 10 9 9 4 10 9 9 9 10 ...
## $ b : Factor w/ 2 levels "f","t": 2 2 2 2 1 2 2 2 2 2 ...
## $ o : Factor w/ 9 levels "a","c","f","l",..: 7 1 4 7 6 1 1 4 7 1 ...
## $ ga : Factor w/ 2 levels "a","f": 2 2 2 2 2 2 2 2 2 2 ...
## $ gs : Factor w/ 2 levels "c","w": 1 1 1 1 2 1 1 1 1 1 ...
## $ gz : Factor w/ 2 levels "b","n": 2 1 1 2 1 1 1 1 2 1 ...
## $ gc : Factor w/ 12 levels "b","e","g","h",..: 5 5 6 6 5 6 3 6 8 3 ...
## $ ss : Factor w/ 2 levels "e","t": 1 1 1 1 2 1 1 1 1 1 ...
## $ sr : Factor w/ 4 levels "b","c","e","r": 3 2 2 3 3 2 2 2 3 2 ...
## $ ssar : Factor w/ 4 levels "f","k","s","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ ssbr : Factor w/ 4 levels "f","k","s","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ scar : Factor w/ 9 levels "b","c","e","g",..: 8 8 8 8 8 8 8 8 8 8 ...
## $ scbr : Factor w/ 9 levels "b","c","e","g",..: 8 8 8 8 8 8 8 8 8 8 ...
## $ vt : Factor w/ 1 level "p": 1 1 1 1 1 1 1 1 1 1 ...
## $ vc : Factor w/ 4 levels "n","o","w","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ rn : Factor w/ 3 levels "n","o","t": 2 2 2 2 2 2 2 2 2 2 ...
## $ rt : Factor w/ 5 levels "e","f","l","n",..: 5 5 5 5 1 5 5 5 5 5 ...
## $ spc : Factor w/ 9 levels "b","h","k","n",..: 3 4 4 3 4 3 3 4 3 3 ...
## $ p : Factor w/ 6 levels "a","c","n","s",..: 4 3 3 4 1 3 3 4 5 4 ...
## $ h : Factor w/ 7 levels "d","g","l","m",..: 6 2 4 6 2 2 4 4 2 4 ...
## $ sr_imp: logi FALSE FALSE FALSE FALSE FALSE FALSE ...mr1 <- sr_imp[c(-17 ,-24)]
str(mr1)## 'data.frame': 8124 obs. of 22 variables:
## $ type: Factor w/ 2 levels "e","p": 2 1 1 2 1 1 1 1 2 1 ...
## $ csh : Factor w/ 6 levels "b","c","f","k",..: 6 6 1 6 6 6 1 1 6 1 ...
## $ csf : Factor w/ 4 levels "f","g","s","y": 3 3 3 4 3 4 3 4 4 3 ...
## $ cc : Factor w/ 10 levels "b","c","e","g",..: 5 10 9 9 4 10 9 9 9 10 ...
## $ b : Factor w/ 2 levels "f","t": 2 2 2 2 1 2 2 2 2 2 ...
## $ o : Factor w/ 9 levels "a","c","f","l",..: 7 1 4 7 6 1 1 4 7 1 ...
## $ ga : Factor w/ 2 levels "a","f": 2 2 2 2 2 2 2 2 2 2 ...
## $ gs : Factor w/ 2 levels "c","w": 1 1 1 1 2 1 1 1 1 1 ...
## $ gz : Factor w/ 2 levels "b","n": 2 1 1 2 1 1 1 1 2 1 ...
## $ gc : Factor w/ 12 levels "b","e","g","h",..: 5 5 6 6 5 6 3 6 8 3 ...
## $ ss : Factor w/ 2 levels "e","t": 1 1 1 1 2 1 1 1 1 1 ...
## $ sr : Factor w/ 4 levels "b","c","e","r": 3 2 2 3 3 2 2 2 3 2 ...
## $ ssar: Factor w/ 4 levels "f","k","s","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ ssbr: Factor w/ 4 levels "f","k","s","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ scar: Factor w/ 9 levels "b","c","e","g",..: 8 8 8 8 8 8 8 8 8 8 ...
## $ scbr: Factor w/ 9 levels "b","c","e","g",..: 8 8 8 8 8 8 8 8 8 8 ...
## $ vc : Factor w/ 4 levels "n","o","w","y": 3 3 3 3 3 3 3 3 3 3 ...
## $ rn : Factor w/ 3 levels "n","o","t": 2 2 2 2 2 2 2 2 2 2 ...
## $ rt : Factor w/ 5 levels "e","f","l","n",..: 5 5 5 5 1 5 5 5 5 5 ...
## $ spc : Factor w/ 9 levels "b","h","k","n",..: 3 4 4 3 4 3 3 4 3 3 ...
## $ p : Factor w/ 6 levels "a","c","n","s",..: 4 3 3 4 1 3 3 4 5 4 ...
## $ h : Factor w/ 7 levels "d","g","l","m",..: 6 2 4 6 2 2 4 4 2 4 ...summary(mr1, maxsum = 12)## type csh csf cc b o ga gs
## e:4208 b: 452 f:2320 b: 168 f:4748 a: 400 a: 210 c:6812
## p:3916 c: 4 g: 4 c: 44 t:3376 c: 192 f:7914 w:1312
## f:3152 s:2556 e:1500 f:2160
## k: 828 y:3244 g:1840 l: 400
## s: 32 n:2284 m: 36
## x:3656 p: 144 n:3528
## r: 16 p: 256
## u: 16 s: 576
## w:1040 y: 576
## y:1072
##
##
## gz gc ss sr ssar ssbr scar scbr
## b:5612 b:1728 e:3516 b:5303 f: 552 f: 600 b: 432 b: 432
## n:2512 e: 96 t:4608 c: 662 k:2372 k:2304 c: 36 c: 36
## g: 752 e:1967 s:5176 s:4936 e: 96 e: 96
## h: 732 r: 192 y: 24 y: 284 g: 576 g: 576
## k: 408 n: 448 n: 512
## n:1048 o: 192 o: 192
## o: 64 p:1872 p:1872
## p:1492 w:4464 w:4384
## r: 24 y: 8 y: 24
## u: 492
## w:1202
## y: 86
## vc rn rt spc p h
## n: 96 n: 36 e:2776 b: 48 a: 384 d:3148
## o: 96 o:7488 f: 48 h:1632 c: 340 g:2148
## w:7924 t: 600 l:1296 k:1872 n: 400 l: 832
## y: 8 n: 36 n:1968 s:1248 m: 292
## p:3968 o: 48 v:4040 p:1144
## r: 72 y:1712 u: 368
## u: 48 w: 192
## w:2388
## y: 48
##
##
## #117 dummy variables
dmy <- dummyVars("~ .", data = mr1, fullRank = TRUE)
#transform to data frame
trsf <- data.frame(predict(dmy, newdata = mr1))
str(trsf[1:10]) #95 variables## 'data.frame': 8124 obs. of 10 variables:
## $ type.p: num 1 0 0 1 0 0 0 0 1 0 ...
## $ csh.c : num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.f : num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.k : num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.s : num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.x : num 1 1 0 1 1 1 0 0 1 0 ...
## $ csf.g : num 0 0 0 0 0 0 0 0 0 0 ...
## $ csf.s : num 1 1 1 0 1 0 1 0 0 1 ...
## $ csf.y : num 0 0 0 1 0 1 0 1 1 0 ...
## $ cc.c : num 0 0 0 0 0 0 0 0 0 0 ...#remove type.p so we can put column back to original
mr2 <- trsf[-1]
str(mr2[1:10])## 'data.frame': 8124 obs. of 10 variables:
## $ csh.c: num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.f: num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.k: num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.s: num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.x: num 1 1 0 1 1 1 0 0 1 0 ...
## $ csf.g: num 0 0 0 0 0 0 0 0 0 0 ...
## $ csf.s: num 1 1 1 0 1 0 1 0 0 1 ...
## $ csf.y: num 0 0 0 1 0 1 0 1 1 0 ...
## $ cc.c : num 0 0 0 0 0 0 0 0 0 0 ...
## $ cc.e : num 0 0 0 0 0 0 0 0 0 0 ...mr2 <- data.frame(mr$type, mr2)
str(mr2[1:10])## 'data.frame': 8124 obs. of 10 variables:
## $ mr.type: Factor w/ 2 levels "e","p": 2 1 1 2 1 1 1 1 2 1 ...
## $ csh.c : num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.f : num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.k : num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.s : num 0 0 0 0 0 0 0 0 0 0 ...
## $ csh.x : num 1 1 0 1 1 1 0 0 1 0 ...
## $ csf.g : num 0 0 0 0 0 0 0 0 0 0 ...
## $ csf.s : num 1 1 1 0 1 0 1 0 0 1 ...
## $ csf.y : num 0 0 0 1 0 1 0 1 1 0 ...
## $ cc.c : num 0 0 0 0 0 0 0 0 0 0 ...names(mr2)[1] <- ("type")
#cleaned set type is factor of e or p
str(mr2[1])## 'data.frame': 8124 obs. of 1 variable:
## $ type: Factor w/ 2 levels "e","p": 2 1 1 2 1 1 1 1 2 1 ...#View(mr2)
#####################################################################################ARTIFICIAL NEURAL NETWORK (ANN)
In this section the data is split 70/30 with random sampling. We confirm the proportions are the same. 51% edible(e) and 48% poisonous (p). The nnet() model is created and tuned with a size of 10 hidden layers, a decay of 0.0001 and a maxit of 500 passes. The values were selected based on other examples found on the internet. The first pass runs 500 and stops at a final value of 0.057. This indicates a good result as it started from 3839. We do a query with varImp() to find the top variables of importance. The top four are sp.c.r, o.c, o.n, gz.n. (spore-print-color-green, odor-creosote, odor-none, gill-size-narrow). Of interest is the other odor categories are in the 5th and 7th of the top ten. odor-foul, and odor-pungent. It is worth mentioning that the spore-print-color-brown and black are also in the top ten. WE run the prediction on the test set and surprisingly come out with an accuracy of 1 and a kappa of 1 which are both the highest metric value. In the table there are no false positives or false negatives. Is this too good to be true or a valid outcome, we will run a cross validation 10 fold to check the results. We run 517 out of 5687 which is about 10% of the random data. The same parameters are used again (size=4, decay=0.0001, maxit=500). The final value is 0.060 (seems good) The result is 1 across ten and a mean of 1. There is confusion here as this should be below 1 but maybe it is valid. A second test is ran with the parameters of(size=5, decay=0.1, and maxit=5000) and came from the “bestTune” parameter in the model, this resulted in A=1 and Kappa=1, the final value was higher at 23.72 so it stopped earlier but not by much. Another try was made with decay=0.0001, maxit=50, size=4 and this was still 100%. It is unknown at this point what else to do to check validation and requires more work to verify the results. The fitted values and residuals are plotted. the fitted are normal 0 and 1. While the residuals are centered around 0, not sure what this means and further time is needed to understand.
######################Neural Net
set.seed(3456)
train_sample <- sample(8124, 5687) #70/30 split
mr_train <- mr2[train_sample, ]
mr_test <- mr2[-train_sample, ]table(mr2$type) #full proportions##
## e p
## 4208 3916prop.table((table(mr_train$type))) #sample proportions##
## e p
## 0.5178477 0.4821523#predict type against everything else
type_model <- nnet(type ~., data = mr_train, size = 10, decay=0.0001, maxit=500)## # weights: 961
## initial value 5129.174670
## iter 10 value 265.027929
## iter 20 value 3.076938
## iter 30 value 0.371356
## iter 40 value 0.352718
## iter 50 value 0.328299
## iter 60 value 0.309906
## iter 70 value 0.296339
## iter 80 value 0.282190
## iter 90 value 0.271781
## iter 100 value 0.254872
## iter 110 value 0.237647
## iter 120 value 0.228582
## iter 130 value 0.219006
## iter 140 value 0.208590
## iter 150 value 0.204293
## iter 160 value 0.197833
## iter 170 value 0.185819
## iter 180 value 0.173900
## iter 190 value 0.168410
## iter 200 value 0.155254
## iter 210 value 0.143915
## iter 220 value 0.132080
## iter 230 value 0.127208
## iter 240 value 0.119249
## iter 250 value 0.115105
## iter 260 value 0.109852
## iter 270 value 0.103697
## iter 280 value 0.099213
## iter 290 value 0.092630
## iter 300 value 0.089279
## iter 310 value 0.086634
## iter 320 value 0.083561
## iter 330 value 0.080222
## iter 340 value 0.076699
## iter 350 value 0.073725
## iter 360 value 0.072143
## iter 370 value 0.070958
## iter 380 value 0.069430
## iter 390 value 0.068156
## iter 400 value 0.066799
## iter 410 value 0.065787
## iter 420 value 0.064691
## iter 430 value 0.063503
## iter 440 value 0.062599
## iter 450 value 0.061279
## iter 460 value 0.060377
## iter 470 value 0.059538
## iter 480 value 0.058777
## iter 490 value 0.058114
## iter 500 value 0.057584
## final value 0.057584
## stopped after 500 iterationsThis ran for 3-4 minutes and this is copied over from the script with results. the last run shows the best model with an accuracy of 0.999 and kappa of 0.9999, a size of 5 and decay of 0.1. The resampling is showing accuracy of 1 and kappa of 1 for all the folds except for 25 where they dropped to A= 0.998 and P=0.996.
cvCtrl <- trainControl(method="repeatedcv", number=10, repeats=3)library(doParallel)## Loading required package: foreach## Loading required package: iterators## Loading required package: parallelcl <- makePSOCKcluster(6)
registerDoParallel(cl)modit = capture.output(train(type ~., data = mr_train,
method="nnet",
preProcess="scale",
trControl=cvCtrl,
maxit=100,
trace=FALSE)
)
stopCluster(cl)plot(table(type_model$fitted.values))
plot(table(type_model$residuals))
###
#sort most influencial variables
topModels <- varImp(type_model)
topModels$Variables <- row.names(topModels)
topModels <- topModels[order(-topModels$Overall),]#sort level of top 10 variables in the model
head(topModels, 10)## Overall Variables
## spc.r 4.860176 spc.r
## o.c 4.213244 o.c
## o.p 3.243726 o.p
## o.n 3.211917 o.n
## gz.n 3.109038 gz.n
## o.f 2.906371 o.f
## spc.n 2.373167 spc.n
## gs.w 2.350670 gs.w
## h.w 2.211128 h.w
## spc.u 2.162081 spc.u#####preds1 <- predict(type_model, newdata = mr_test, type="class")
table(preds1, mr_test$type)##
## preds1 e p
## e 1263 0
## p 0 1174head(preds1, 10)## [1] "p" "e" "e" "p" "e" "p" "p" "e" "e" "e"postResample(preds1, mr_test$type) #accuracy and kappa## Accuracy Kappa
## 1 1totalError <- c()
#split data by 10 portions
cv <- 10
cvDivider <- floor(nrow(mr_train) / (cv+1))
cvDivider #number of rows## [1] 517nrow(mr_train) #roughly 10x the row## [1] 5687# loop data on 10 different sections of the data increase by a 10th each run, put in best tune parameters
capture.output(
for (cv in seq(1:cv)) {
#assign chunk to data set
dataTestIndex <- c((cv * cvDivider):(cv * cvDivider + cvDivider))
dataTest <- mr_train[dataTestIndex, ]
#everything else to train
dataTrain <- mr_train[-dataTestIndex, ]
type_model <- nnet(type~., data = mr_train, size = 5, decay=0.1, maxit=500)
pred <- predict(type_model, dataTest)
#classification error
err <- ce(as.numeric(dataTest$type), as.numeric(pred))
totalError <- c(totalError, err)
}
)## [1] "# weights: 481" "initial value 4131.038209 "
## [3] "iter 10 value 455.101632" "iter 20 value 214.076274"
## [5] "iter 30 value 117.864798" "iter 40 value 70.556964"
## [7] "iter 50 value 42.605632" "iter 60 value 31.430671"
## [9] "iter 70 value 26.466643" "iter 80 value 25.158454"
## [11] "iter 90 value 24.501575" "iter 100 value 24.213052"
## [13] "iter 110 value 24.127779" "iter 120 value 24.021257"
## [15] "iter 130 value 23.970028" "iter 140 value 23.939620"
## [17] "iter 150 value 23.907873" "iter 160 value 23.890786"
## [19] "iter 170 value 23.882737" "iter 180 value 23.870609"
## [21] "iter 190 value 23.858447" "iter 200 value 23.847741"
## [23] "iter 210 value 23.833776" "iter 220 value 23.830237"
## [25] "iter 230 value 23.829832" "iter 240 value 23.829748"
## [27] "iter 250 value 23.829601" "iter 260 value 23.829033"
## [29] "iter 270 value 23.828754" "iter 280 value 23.828671"
## [31] "iter 290 value 23.828635" "final value 23.828632 "
## [33] "converged" "# weights: 481"
## [35] "initial value 3903.043295 " "iter 10 value 516.137283"
## [37] "iter 20 value 281.185677" "iter 30 value 130.188377"
## [39] "iter 40 value 64.177186" "iter 50 value 38.173988"
## [41] "iter 60 value 31.025100" "iter 70 value 27.980851"
## [43] "iter 80 value 26.268903" "iter 90 value 25.322536"
## [45] "iter 100 value 24.785172" "iter 110 value 24.384057"
## [47] "iter 120 value 24.280135" "iter 130 value 24.179441"
## [49] "iter 140 value 24.062253" "iter 150 value 23.926230"
## [51] "iter 160 value 23.889262" "iter 170 value 23.878509"
## [53] "iter 180 value 23.874436" "iter 190 value 23.870764"
## [55] "iter 200 value 23.866497" "iter 210 value 23.862410"
## [57] "iter 220 value 23.860302" "iter 230 value 23.853558"
## [59] "iter 240 value 23.836053" "iter 250 value 23.830244"
## [61] "iter 260 value 23.828885" "iter 270 value 23.828658"
## [63] "iter 280 value 23.828641" "iter 290 value 23.828635"
## [65] "final value 23.828632 " "converged"
## [67] "# weights: 481" "initial value 4463.836252 "
## [69] "iter 10 value 305.131771" "iter 20 value 85.451644"
## [71] "iter 30 value 52.717735" "iter 40 value 40.201806"
## [73] "iter 50 value 34.720639" "iter 60 value 33.113350"
## [75] "iter 70 value 30.495137" "iter 80 value 27.741541"
## [77] "iter 90 value 26.456015" "iter 100 value 25.899196"
## [79] "iter 110 value 25.724112" "iter 120 value 25.582297"
## [81] "iter 130 value 25.512619" "iter 140 value 25.220500"
## [83] "iter 150 value 24.859076" "iter 160 value 24.393029"
## [85] "iter 170 value 24.078532" "iter 180 value 24.025912"
## [87] "iter 190 value 24.001804" "iter 200 value 23.972064"
## [89] "iter 210 value 23.907833" "iter 220 value 23.881270"
## [91] "iter 230 value 23.852015" "iter 240 value 23.830189"
## [93] "iter 250 value 23.828632" "iter 250 value 23.828632"
## [95] "iter 250 value 23.828632" "final value 23.828632 "
## [97] "converged" "# weights: 481"
## [99] "initial value 3922.557831 " "iter 10 value 198.221549"
## [101] "iter 20 value 54.769153" "iter 30 value 31.934519"
## [103] "iter 40 value 27.472269" "iter 50 value 25.915732"
## [105] "iter 60 value 25.049463" "iter 70 value 24.456231"
## [107] "iter 80 value 23.992637" "iter 90 value 23.903161"
## [109] "iter 100 value 23.882183" "iter 110 value 23.863807"
## [111] "iter 120 value 23.857866" "iter 130 value 23.852526"
## [113] "iter 140 value 23.851162" "iter 150 value 23.850689"
## [115] "iter 160 value 23.849050" "iter 170 value 23.846435"
## [117] "iter 180 value 23.845368" "iter 190 value 23.845244"
## [119] "final value 23.845145 " "converged"
## [121] "# weights: 481" "initial value 4108.098129 "
## [123] "iter 10 value 77.935245" "iter 20 value 35.428331"
## [125] "iter 30 value 30.075878" "iter 40 value 27.549116"
## [127] "iter 50 value 26.587039" "iter 60 value 25.595512"
## [129] "iter 70 value 24.886336" "iter 80 value 24.211819"
## [131] "iter 90 value 24.115509" "iter 100 value 24.052371"
## [133] "iter 110 value 23.966428" "iter 120 value 23.945243"
## [135] "iter 130 value 23.904446" "iter 140 value 23.896764"
## [137] "iter 150 value 23.876036" "iter 160 value 23.860088"
## [139] "iter 170 value 23.843901" "iter 180 value 23.835529"
## [141] "iter 190 value 23.830551" "final value 23.828632 "
## [143] "converged" "# weights: 481"
## [145] "initial value 4133.554002 " "iter 10 value 428.491443"
## [147] "iter 20 value 202.828516" "iter 30 value 68.434375"
## [149] "iter 40 value 49.453764" "iter 50 value 42.202017"
## [151] "iter 60 value 39.013840" "iter 70 value 34.744057"
## [153] "iter 80 value 31.933707" "iter 90 value 29.732941"
## [155] "iter 100 value 28.384519" "iter 110 value 27.048713"
## [157] "iter 120 value 26.412491" "iter 130 value 26.142390"
## [159] "iter 140 value 25.808631" "iter 150 value 25.485091"
## [161] "iter 160 value 25.415397" "iter 170 value 25.312302"
## [163] "iter 180 value 25.234511" "iter 190 value 24.986085"
## [165] "iter 200 value 24.403117" "iter 210 value 24.090090"
## [167] "iter 220 value 23.914432" "iter 230 value 23.851080"
## [169] "iter 240 value 23.843012" "iter 250 value 23.838547"
## [171] "iter 260 value 23.833956" "iter 270 value 23.831865"
## [173] "iter 280 value 23.828783" "iter 290 value 23.828708"
## [175] "iter 300 value 23.828660" "iter 310 value 23.828633"
## [177] "final value 23.828632 " "converged"
## [179] "# weights: 481" "initial value 4166.413738 "
## [181] "iter 10 value 576.127011" "iter 20 value 202.529239"
## [183] "iter 30 value 61.682299" "iter 40 value 33.779033"
## [185] "iter 50 value 27.813710" "iter 60 value 26.142641"
## [187] "iter 70 value 24.935432" "iter 80 value 24.730898"
## [189] "iter 90 value 24.668778" "iter 100 value 24.594535"
## [191] "iter 110 value 24.553234" "iter 120 value 24.549415"
## [193] "iter 130 value 24.547717" "iter 140 value 24.547243"
## [195] "final value 24.547226 " "converged"
## [197] "# weights: 481" "initial value 4465.179580 "
## [199] "iter 10 value 456.875594" "iter 20 value 213.634932"
## [201] "iter 30 value 150.275506" "iter 40 value 92.840494"
## [203] "iter 50 value 38.199064" "iter 60 value 29.177637"
## [205] "iter 70 value 27.175464" "iter 80 value 25.607727"
## [207] "iter 90 value 25.084173" "iter 100 value 24.742807"
## [209] "iter 110 value 24.619621" "iter 120 value 24.521725"
## [211] "iter 130 value 24.433243" "iter 140 value 24.158239"
## [213] "iter 150 value 24.026866" "iter 160 value 23.990799"
## [215] "iter 170 value 23.979853" "iter 180 value 23.960466"
## [217] "iter 190 value 23.932998" "iter 200 value 23.892964"
## [219] "iter 210 value 23.859856" "iter 220 value 23.846410"
## [221] "iter 230 value 23.836721" "iter 240 value 23.831773"
## [223] "iter 250 value 23.830133" "iter 260 value 23.829567"
## [225] "iter 270 value 23.829192" "iter 280 value 23.829040"
## [227] "iter 290 value 23.828738" "iter 300 value 23.828635"
## [229] "final value 23.828634 " "converged"
## [231] "# weights: 481" "initial value 4371.291868 "
## [233] "iter 10 value 780.885653" "iter 20 value 587.825721"
## [235] "iter 30 value 363.720253" "iter 40 value 139.595418"
## [237] "iter 50 value 58.480965" "iter 60 value 38.726359"
## [239] "iter 70 value 30.458288" "iter 80 value 26.274315"
## [241] "iter 90 value 24.739274" "iter 100 value 24.222307"
## [243] "iter 110 value 24.045235" "iter 120 value 23.971757"
## [245] "iter 130 value 23.920575" "iter 140 value 23.879082"
## [247] "iter 150 value 23.867271" "iter 160 value 23.857194"
## [249] "iter 170 value 23.852346" "iter 180 value 23.850086"
## [251] "iter 190 value 23.848909" "iter 200 value 23.848579"
## [253] "iter 210 value 23.848370" "iter 220 value 23.848232"
## [255] "iter 230 value 23.848019" "iter 240 value 23.847315"
## [257] "iter 250 value 23.845781" "iter 260 value 23.845296"
## [259] "iter 270 value 23.845176" "iter 280 value 23.845153"
## [261] "iter 290 value 23.845146" "final value 23.845146 "
## [263] "converged" "# weights: 481"
## [265] "initial value 3976.505403 " "iter 10 value 479.875071"
## [267] "iter 20 value 399.158322" "iter 30 value 246.440189"
## [269] "iter 40 value 131.962324" "iter 50 value 63.162088"
## [271] "iter 60 value 41.803408" "iter 70 value 31.054774"
## [273] "iter 80 value 26.723853" "iter 90 value 24.936619"
## [275] "iter 100 value 24.013096" "iter 110 value 23.924894"
## [277] "iter 120 value 23.873463" "iter 130 value 23.850006"
## [279] "iter 140 value 23.841252" "iter 150 value 23.838300"
## [281] "iter 160 value 23.837006" "iter 170 value 23.835772"
## [283] "iter 180 value 23.834722" "iter 190 value 23.833913"
## [285] "iter 200 value 23.832358" "iter 210 value 23.830529"
## [287] "iter 220 value 23.829161" "iter 230 value 23.829024"
## [289] "iter 240 value 23.828997" "iter 250 value 23.828963"
## [291] "iter 260 value 23.828898" "iter 270 value 23.828748"
## [293] "iter 280 value 23.828684" "iter 290 value 23.828650"
## [295] "iter 300 value 23.828636" "final value 23.828633 "
## [297] "converged"totalError## [1] 1 1 1 1 1 1 1 1 1 1mean(totalError)## [1] 1# run code with 50 iterations
capture.output(
for (cv in seq(1:cv)) {
#assign chunk to data set
dataTestIndex <- c((cv * cvDivider):(cv * cvDivider + cvDivider))
dataTest <- mr_train[dataTestIndex, ]
#everything else to train
dataTrain <- mr_train[-dataTestIndex, ]
type_model <- nnet(type~., data = mr_train, size = 4, decay=0.0001, maxit=50)
pred <- predict(type_model, dataTest)
#classification error
err <- ce(as.numeric(dataTest$type), as.numeric(pred))
totalError <- c(totalError, err)
}
)## [1] "# weights: 385" "initial value 3992.215378 "
## [3] "iter 10 value 225.688621" "iter 20 value 14.815525"
## [5] "iter 30 value 0.631761" "iter 40 value 0.229973"
## [7] "iter 50 value 0.221208" "final value 0.221208 "
## [9] "stopped after 50 iterations" "# weights: 385"
## [11] "initial value 3919.352988 " "iter 10 value 230.359279"
## [13] "iter 20 value 13.853211" "iter 30 value 0.319568"
## [15] "iter 40 value 0.232561" "iter 50 value 0.221478"
## [17] "final value 0.221478 " "stopped after 50 iterations"
## [19] "# weights: 385" "initial value 3800.270363 "
## [21] "iter 10 value 985.054588" "iter 20 value 22.792292"
## [23] "iter 30 value 1.071623" "iter 40 value 0.992320"
## [25] "iter 50 value 0.913452" "final value 0.913452 "
## [27] "stopped after 50 iterations" "# weights: 385"
## [29] "initial value 4287.392108 " "iter 10 value 343.656550"
## [31] "iter 20 value 26.949759" "iter 30 value 1.446327"
## [33] "iter 40 value 0.804258" "iter 50 value 0.680953"
## [35] "final value 0.680953 " "stopped after 50 iterations"
## [37] "# weights: 385" "initial value 4384.903759 "
## [39] "iter 10 value 532.228491" "iter 20 value 37.648244"
## [41] "iter 30 value 37.373940" "iter 40 value 37.314777"
## [43] "iter 50 value 37.276753" "final value 37.276753 "
## [45] "stopped after 50 iterations" "# weights: 385"
## [47] "initial value 3798.281114 " "iter 10 value 634.725052"
## [49] "iter 20 value 277.357662" "iter 30 value 274.777653"
## [51] "iter 40 value 274.727454" "iter 50 value 274.713995"
## [53] "final value 274.713995 " "stopped after 50 iterations"
## [55] "# weights: 385" "initial value 3891.212678 "
## [57] "iter 10 value 470.883388" "iter 20 value 14.524364"
## [59] "iter 30 value 0.373357" "iter 40 value 0.328683"
## [61] "iter 50 value 0.305251" "final value 0.305251 "
## [63] "stopped after 50 iterations" "# weights: 385"
## [65] "initial value 4576.628512 " "iter 10 value 839.380735"
## [67] "iter 20 value 186.731047" "iter 30 value 17.636580"
## [69] "iter 40 value 1.326494" "iter 50 value 1.093621"
## [71] "final value 1.093621 " "stopped after 50 iterations"
## [73] "# weights: 385" "initial value 3973.229118 "
## [75] "iter 10 value 299.981309" "iter 20 value 119.312181"
## [77] "iter 30 value 9.965782" "iter 40 value 0.337041"
## [79] "iter 50 value 0.271618" "final value 0.271618 "
## [81] "stopped after 50 iterations" "# weights: 385"
## [83] "initial value 4325.988622 " "iter 10 value 393.708448"
## [85] "iter 20 value 10.199643" "iter 30 value 0.148243"
## [87] "iter 40 value 0.123917" "iter 50 value 0.113179"
## [89] "final value 0.113179 " "stopped after 50 iterations"totalError## [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1mean(totalError) #compare to error previously## [1] 1SUPPORT VECTOR MACHINE (SVM)
This next section is going to use the same data and same training and test sets used above for the ANN. The first svm model using the “vanilladot” kernel produces a similar result to the ANN. The training error is 0, meaning this is 100% accurate. The second kernel is the rbfdot and produces error of 0.000352.For the rbfdot the table is almost exactly the same for the ANN but with 1 false negative slipping its way in to the model. 1276 TP/ 1160 TN. Where the ANN had 1277 TP and 1161 TN. The third kernel is the polydot and it too produces an error of 0]. At this point we don’t know why as it is expected to have different outcomes for each kernel and this is not the case. Are there some errors somewhere not being detected or is this in fact a valid out come. We will run a 10 fold cross validation to to see what is going on with the model. The CART output shows the correct 5687 samples with 94 predictors and the tow classes (e) and (p). The 10-fold cross validation was run 5 times and the cp0.044 model had the highest accuracy of 0.957 and kappa of 0.91. These seem very reasonable and less than a perfect 1. Of interest for comparison is the varImp which gives 9 with an overall and the remaining with 0. We see Odor-neutral at the top with ring-type-pendant, odor-foul, and two stalk-surface variables and stalk-root-club showing value for this model. The good thing is there is some similarity between ANN and SVM with o.n, o.f, ssar.k, gz.n, and sp.c.r all were important in the model for both ANN and SVM. At this point maybe it could help to reduce the variables down to the top 10 combined from both models and rerun on those to see if there is a difference or run on a different set of data that the model hasn’t seen to predict if it is in fact highly accurate.
#SVM
library(e1071)
library(kernlab)set.seed(6789)
mr_classifier <- ksvm(type ~., data = mr_train, kernel = "vanilladot")## Setting default kernel parametersmr_classifier## Support Vector Machine object of class "ksvm"
##
## SV type: C-svc (classification)
## parameter : cost C = 1
##
## Linear (vanilla) kernel function.
##
## Number of Support Vectors : 161
##
## Objective Function Value : -0.4639
## Training error : 0mr_predictions <- predict(mr_classifier, mr_test)
head(mr_predictions)## [1] p e e p e p
## Levels: e ptable(mr_predictions, mr_test$type)##
## mr_predictions e p
## e 1263 0
## p 0 1174agreement <- mr_predictions == mr_test$type
table(agreement)## agreement
## TRUE
## 2437prop.table(table(agreement))## agreement
## TRUE
## 1#rbfdot kernal
mr_classifier_rbfdot <- ksvm(type ~., data = mr_train, kernel = "rbfdot")
mr_classifier_rbfdot## Support Vector Machine object of class "ksvm"
##
## SV type: C-svc (classification)
## parameter : cost C = 1
##
## Gaussian Radial Basis kernel function.
## Hyperparameter : sigma = 0.00850323450395857
##
## Number of Support Vectors : 880
##
## Objective Function Value : -46.1997
## Training error : 0.000176mr_predictions__rbfdot <- predict(mr_classifier_rbfdot, mr_test)
head(mr_predictions__rbfdot)## [1] p e e p e p
## Levels: e ptable(mr_predictions__rbfdot, mr_test$type)##
## mr_predictions__rbfdot e p
## e 1263 3
## p 0 1171agreement_rbfdot <- mr_predictions__rbfdot == mr_test$type
table(agreement_rbfdot)## agreement_rbfdot
## FALSE TRUE
## 3 2434prop.table(table(agreement_rbfdot))## agreement_rbfdot
## FALSE TRUE
## 0.001231022 0.998768978# polydot kernal
mr_classifier_poly <- ksvm(type ~., data = mr_train, kernel = "polydot")## Setting default kernel parametersmr_classifier_poly## Support Vector Machine object of class "ksvm"
##
## SV type: C-svc (classification)
## parameter : cost C = 1
##
## Polynomial kernel function.
## Hyperparameters : degree = 1 scale = 1 offset = 1
##
## Number of Support Vectors : 202
##
## Objective Function Value : -0.4639
## Training error : 0mr_predictions_poly <- predict(mr_classifier_poly, mr_test)
head(mr_predictions_poly)## [1] p e e p e p
## Levels: e ptable(mr_predictions_poly, mr_test$type)##
## mr_predictions_poly e p
## e 1263 0
## p 0 1174agreement_poly <- mr_predictions_poly == mr_test$type
table(agreement_poly)## agreement_poly
## TRUE
## 2437prop.table(table(agreement_poly))## agreement_poly
## TRUE
## 1# SVM Cross Validate
control <- trainControl(method="repeatedcv", number=10, repeats = 5)set.seed(7495)
cl <- makePSOCKcluster(6)
registerDoParallel(cl)
modelsvm <- train(type ~.,
data = mr_train,
method="rpart",
preProcess="scale",
trControl=control)
stopCluster(cl)modelsvm## CART
##
## 5687 samples
## 94 predictor
## 2 classes: 'e', 'p'
##
## Pre-processing: scaled (94)
## Resampling: Cross-Validated (10 fold, repeated 5 times)
## Summary of sample sizes: 5118, 5118, 5118, 5118, 5119, 5119, ...
## Resampling results across tuning parameters:
##
## cp Accuracy Kappa
## 0.05069292 0.9542847 0.9085469
## 0.12472648 0.9105378 0.8217678
## 0.76002918 0.6950380 0.3729620
##
## Accuracy was used to select the optimal model using the largest value.
## The final value used for the model was cp = 0.05069292.importance <- varImp(modelsvm, scale = FALSE)impM = data.frame(importance$importance)
impM$Vars = row.names(impM)
impM[order(-impM$Overall),][1:10,]## Overall Vars
## o.n 1736.2559 o.n
## rt.p 1221.7021 rt.p
## o.f 1096.6735 o.f
## ssar.k 958.4037 ssar.k
## ssbr.k 908.2624 ssbr.k
## sr.c 511.6481 sr.c
## b.t 484.0714 b.t
## o.l 431.1686 o.l
## h.m 252.5091 h.m
## csh.c 0.0000 csh.cplot(varImp(modelsvm), top =10)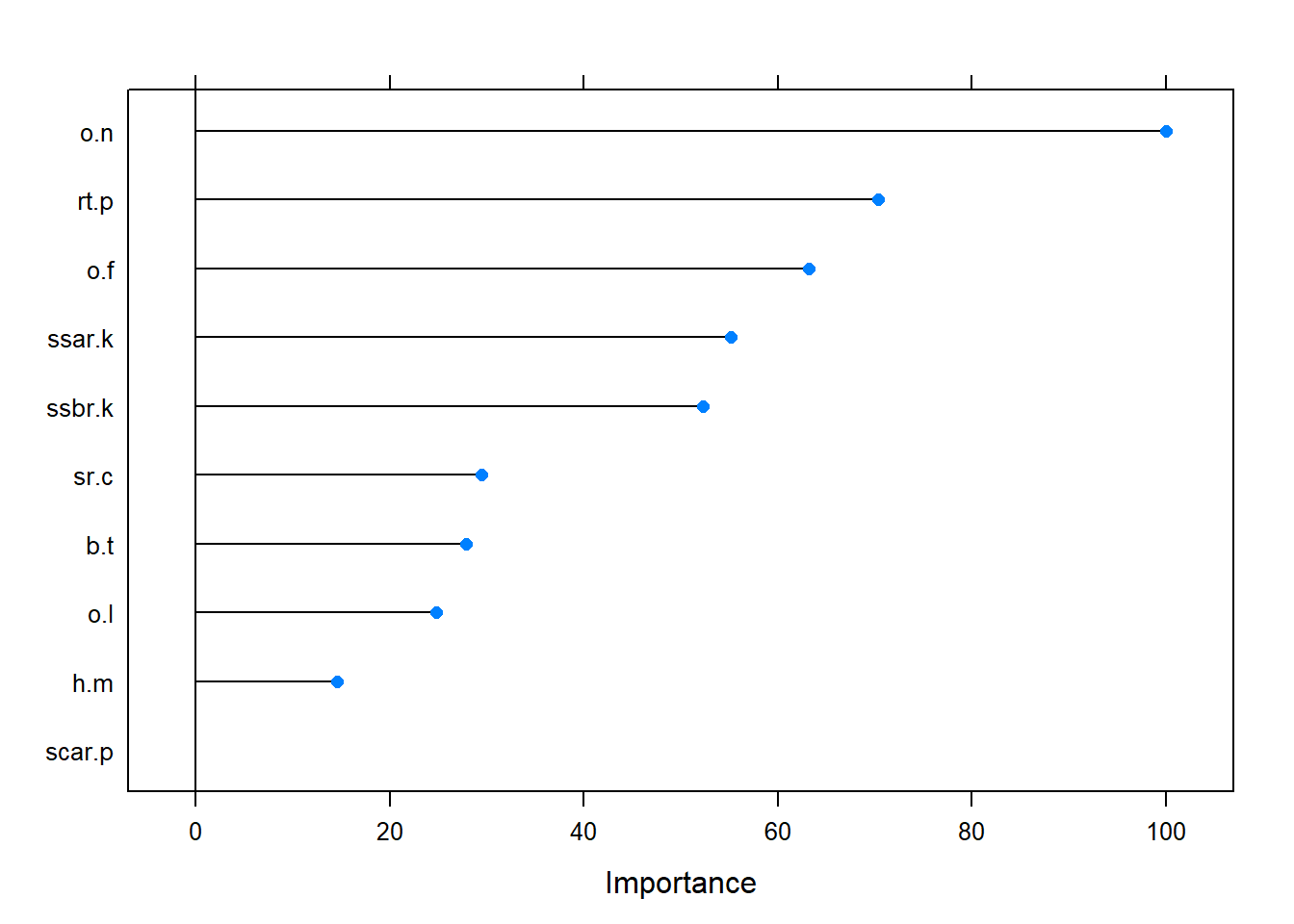
library(rpart)model.rp <- rpart(type ~., data = mr_train)
model.rp$variable.importance## o.n gs.w sr.c o.f gz.n ssar.k rt.p
## 1736.25595 513.92930 511.64807 499.62666 464.09134 425.00248 415.76329
## h.m sr.r ssbr.y p.n o.l spc.r b.t
## 280.04675 250.05114 250.05114 228.27376 179.99964 95.67448 73.98649
## o.c ss.t gc.k spc.k csh.f csh.x o.m
## 73.98649 73.98649 65.59591 39.52703 35.47297 35.47297 31.48604
## gc.r spc.u
## 24.87537 17.48186